Zygomycota
Microscopic 'Pin' or 'Sugar' Molds
Timothy Y. James and Kerry O'Donnell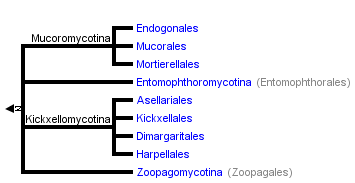

The tree shown above is based on several phylogenetic analyses of gene sequences of nuclear small subunit (SSU) ribosomal DNA (Lutzoni et al. 2004, Nagahama et al. 1995, O'Donnell et al. 1998, Tanabe et al. 2000), elongation factor 1α, the largest subunit of RNA polymerase II (RPB1; Tanabe et al. 2004), and combined analysis of multiple genes (James et al. 2006; White et al. 2006; Liu et al. 2006). Most of the clades shown are strongly supported as monophyletic, but relationships among them are poorly resolved. Moreover, monophyly of the Zygomycota remains controversial, and it was not included as a formal taxon in the "AFTOL classification" of Fungi (Hibbett et al., 2007). This page is currently being revised to reflect understanding of the phylogeny of taxa formerly placed in Zygomycota.
Containing group: Fungi
Introduction
The Zygomycota contains approximately 1% of the described species of true Fungi (~900 described species; Kirk et al. 2001). The most familiar representatives include the fast-growing molds that we encounter on spoiled strawberries (Figure 1) and other fruits high in sugar content. Although these fungi are common in terrestrial and aquatic ecosystems, they are rarely noticed by humans because they are of microscopic size. Colonial growth and the taxonomically informative asexual reproductive structures Zygomycota produce are typically studied after culturing on various agar media. Direct microscopic observation of suitable substrates is required for those species that either have not or cannot be cultured. Fewer than half of the species have been cultured and the majority of these are members of the Mucorales, a group that includes some of the fastest growing fungi.Zygomycota are defined and distinguished from all other fungi by sexual reproduction via zygospores following gametangial fusion (Figure 2A,B) and asexual reproduction by uni-to-multispored sporangia (Figure 3A,B) within which nonmotile, single-celled sporangiospores are produced. The phylum comprises at least seven phylogenetically diverse orders. Monophyly of the phylum and interrelationships among orders are currently under intensive investigation using multilocus DNA sequence data. This introduction to the Zygomycota does not include the order comprising the arbuscular mycorrhizal (AM) fungi, the Glomales, because it has been elevated to the rank of phylum, the Glomeromycota (Schüßler et al. 2001). One other group, the Microsporidia, were previously considered protozoa, however, DNA, biochemistry, and morphology suggest these highly reduced, obligate, intracellular parasites may have evolved from a zygomycete-like ancestor (Keeling 2003).

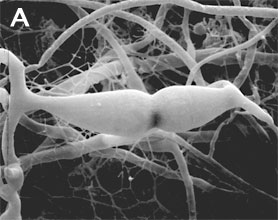
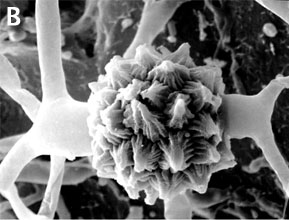 Figure 2. Sexual reproduction. (A) Scanning electron micrograph of gametangial fusion in Mucor mucedo. (B) Highly ornamented zygosporangium of Mycotypha africana. (From O'Donnell 1979).
Figure 2. Sexual reproduction. (A) Scanning electron micrograph of gametangial fusion in Mucor mucedo. (B) Highly ornamented zygosporangium of Mycotypha africana. (From O'Donnell 1979).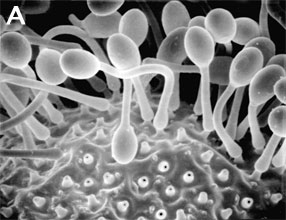
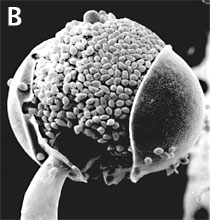 Figure 3. Asexual reproduction. (A) Scanning electron micrograph of unispored sporangia of Benjaminiella poitrasii and (B) dehisced multispored sporangium of Gilbertella persicaria releasing sporangiospores. (From O'Donnell 1979).
Figure 3. Asexual reproduction. (A) Scanning electron micrograph of unispored sporangia of Benjaminiella poitrasii and (B) dehisced multispored sporangium of Gilbertella persicaria releasing sporangiospores. (From O'Donnell 1979).Conversely, some species have a negative economic impact on human affairs by causing storage rots of fruits (particularly strawberries by Rhizopus stolonifer [Figure 1]), as agents of plant disease (e.g., Choanephora cucurbitarum flower rot of curcurbits), while other species can cause life-threatening opportunistic infections of diabetic, immuno-suppressed, and immuno-compromised patients (de Hoog et al. 2000). In addition to some Mucorales that attack immuno-suppressed humans, several species of microsporidia cause serious human infections. Some zygomycetes are regularly isolated by veterinarians from domesticated animals in tropical and subtropical regions of the world, including the US gulf states.
Characteristics
Zygomycota, like all true fungi, produce cell walls containing chitin. They grow primarily as mycelia, or filaments of long cells called hyphae. Unlike the so-called 'higher fungi' comprising the Ascomycota and Basidiomycota which produce regularly septate mycelia, most Zygomycota form hyphae which are generally coenocytic because they lack cross walls or septa. There are, however, several exceptions and septa may form at irregular intervals throughout the older parts of the mycelium or are regularly spaced in two sister orders of Zygomycota, the Kickxellales and Harpellales.The unique character (synapomorphy) of the Zygomycota is the zygospore. Zygospores are formed within a zygosporangium after the fusion of specialized hyphae called gametangia during the sexual cycle (Figure 2A). A single zygospore is formed per zygosporangium. Because of this one-to-one relationship, the terms are often used interchangably. The mature zygospore is often thick-walled (Figure 2B), and undergoes an obligatory dormant period before germination. Most Zygomycota are thought to have a zygotic or haplontic life cycle (Figure 4). Thus, the only diploid phase takes place within the zygospore. Nuclei within the zygospore are believed to undergo meiosis during germination, but this has only been demonstrated genetically within the model eukaryote Phycomyces blakesleeanus (Eslava et al. 1975).

 Figure 4. Generalized life cycle of Zygomycota. Asexual reproduction occurs primarily by sporangiospores produced by mitosis and cell division. The only diploid (2N) phase in the life cycle is the zygospore, produced through the conjugation of compatible gametangia during the sexual cycle (see Figure 2A, B).
Figure 4. Generalized life cycle of Zygomycota. Asexual reproduction occurs primarily by sporangiospores produced by mitosis and cell division. The only diploid (2N) phase in the life cycle is the zygospore, produced through the conjugation of compatible gametangia during the sexual cycle (see Figure 2A, B).Sporangia are formed at the ends of specialized hyphae called sporangiophores. In the model organism, Phycomyces blakesleeanus, sporangial development has been studied extensively to understand the genetic basis for various trophisms, including the strong phototrophic responses to blue light. A unique spore dispersal strategy for the Mucorales is exhibited by the dung fungus Pilobolus, whose name literally means 'the hat thrower' (see far left Title illustration). The entire black sporangium is explosively shot off of the top of the sporangiophore up to distances of several meters. Phototrophic growth of the sporangiophore facilitates dispersal away from the dung onto a fresh blade of grass where it may be consumed by an herbivore, thereby completing the asexual cycle after the spores pass through the digestive system. Some members of the Entomophthorales (e.g., Basidiobolus, Conidiobolus) also reproduce via forcibly discharged asexual spores. Interestingly, species of Basidiobolus, Conidiobolus and several other genera produce a second kind of spore on a long stalk that appears to have certain morphological adaptations for efficient insect dispersal.

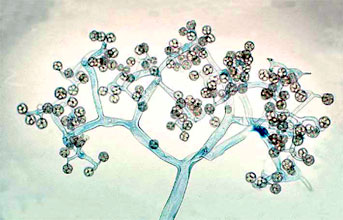 Figure 5. Dichotomously branching sporangiophore of Thamnidium elegans (Mucorales). The few-spored sporangiola are borne at the tips of the sporangiophore branches (© G. L. Barron 2004).
Figure 5. Dichotomously branching sporangiophore of Thamnidium elegans (Mucorales). The few-spored sporangiola are borne at the tips of the sporangiophore branches (© G. L. Barron 2004).
 Figure 6. Scanning electron micrograph of uniseriate merosporangia produced on a vesicle (hidden beneath merosporangia) of Syncephalastrum racemosum (Mucorales). (From O'Donnell 1979).
Figure 6. Scanning electron micrograph of uniseriate merosporangia produced on a vesicle (hidden beneath merosporangia) of Syncephalastrum racemosum (Mucorales). (From O'Donnell 1979).
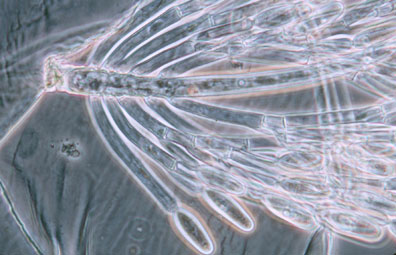 Figure 7. Photo of thallus of Genistellospora homothallica (Harpellales) bearing trichospores attached to the hindgut cuticle of a Chilean blackfly. (© Misra and Lichtwardt 2000).
Figure 7. Photo of thallus of Genistellospora homothallica (Harpellales) bearing trichospores attached to the hindgut cuticle of a Chilean blackfly. (© Misra and Lichtwardt 2000).Zygomycota also participate in a number of interesting symbioses. As mentioned above, the Harpellales inhabit arthropods (particularly freshwater aquatic insect larvae; Figure 7) where they are attached to the chitinous lining of the hindgut. Harpellids presumably feed on nutrients that are not utilized by the arthropod. Because they are generally assumed to neither harm nor benefit the host animal, this association is considered commensalistic. In contrast, the Entomophthorales include many insect pathogens that can cause huge disease outbreaks (see center Title slide showing infected maggot fly). Some of this pathogenicity is being tapped for use in the biocontrol of specific insect pests, including periodical cicadas (Bidochka et al. 1996; Hajek 1999). A number of other Zygomycota are mycoparasitic, or parasites of other fungi. All members of the Dimargaritales (only 15 species) and many Zoopagales are typically obligate parasites of mucoralean hosts. Other mycoparasites in the Mucorales (e.g., Syzygites, Spinellus) specialize on mushroom fruiting bodies (Basidiomycota; Figure 8).

 Figure 8. Sporangia of Spinellus fusiger (Mucorales) parasitic on fruitbodies of the mushroom Mycena pura. (© Malcolm Storey 2004).
Figure 8. Sporangia of Spinellus fusiger (Mucorales) parasitic on fruitbodies of the mushroom Mycena pura. (© Malcolm Storey 2004).
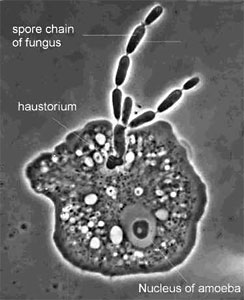 Figure 9. The parasite Amoebophilus simplex (Zoopagales) and its amoeba host. Nutrient transfer occurs through a specialized hypha called a haustorium that enters the amoeba. Spores are produced in chains, and they are engulfed by scavenging amoebae to begin the infection process (© G. L. Barron 2004).
Figure 9. The parasite Amoebophilus simplex (Zoopagales) and its amoeba host. Nutrient transfer occurs through a specialized hypha called a haustorium that enters the amoeba. Spores are produced in chains, and they are engulfed by scavenging amoebae to begin the infection process (© G. L. Barron 2004).Discussion of Phylogenetic Relationships
The Zygomycota are thought to have diverged from the remaining fungi before the colonization of land by plants 600-1,400 million years ago (Berbee and Taylor 2001; Heckman et al. 2001). Molecular phylogenetic studies place the Zygomycota near the base of the kingdom Fungi, diverging after the Chytridiomycota, the most basal fungal lineage (James et al. 2006; White et al. 2006). However, as presently circumscribed, it is uncertain whether the Zygomycota represent a monophyletic group. Studies using SSU rDNA sequence data have generated molecular phylogenies suggesting the Zygomycota may be either para- or polyphyletic (Bruns et al. 1992; Tanabe et al. 2000, 2004). With the recent removal of the Glomales from the Zygomycota (Schüßler et al. 2001), this phylum is restricted to species which form zygospores through mycelial conjugation, at least in those species where sexual reproduction is known.Prior to the use of molecular phylogenetics, the Zygomycota were classified into two classes, the Zygomycetes and Trichomycetes (Alexopoulos et al. 1996). Analyses of SSU rDNA sequences, however, have shown that the Trichomycetes are polyphyletic, comprising what we now know are Ichthyosporean protozoans related to animals (Benny and O'Donnell 2000; Ustinova et al. 2000) and also some true Fungi, the Harpellales, which are nested within the Zygomycetes (O'Donnell et al. 1998; Tanabe et al. 2000). Although relationships among the orders are poorly understood, analyses of RPB1 DNA sequences resolved a clade comprising the Kickxellales-Harpellales-Dimargaritales (Tanabe et al. 2004). A morphological synapomorphy for this clade is the possession of a uniperforate septum with a lenticular cavity (Figure 10; Benny et al. 2001). A large-scale phylogeny of the Mucorales, using three genes and at least one member of each recognized genus, suggests that several of the largest families and the two largest genera (Mucor and Absidia) are polyphyletic (O'Donnell et al. 2001).

 Figure 10. Transmission electron micrograph of vegetative hypha of Kickxella alabastrina (Kickxellales). White line separating the upper from the lower cell is a section of the cross wall or septum. Note the lens shaped plug that lies within the septal pore (lenticular cavity). Scale bar = 0.5 µm. (From Tanabe et al. 2004; © Elsevier 2003).
Figure 10. Transmission electron micrograph of vegetative hypha of Kickxella alabastrina (Kickxellales). White line separating the upper from the lower cell is a section of the cross wall or septum. Note the lens shaped plug that lies within the septal pore (lenticular cavity). Scale bar = 0.5 µm. (From Tanabe et al. 2004; © Elsevier 2003).Though controversial, congruent evidence from alpha- and beta-tubulin gene phylogenies support a zygomycete origin of the microsporidia, a group of highly reduced obligate intracellular parasites of a wide variety of animals including humans (Keeling et al. 2000; Keeling 2003). Because several microsporidian species have emerged as major pathogens of immuno-compromised patients over the past two decades, this enigmatic group has received considerable attention recently by the scientific community. Placement of the microsporidia, however, remains controversial.
 05.42
05.42
 mizukage
mizukage




















0 komentar:
Posting Komentar